- Introduction
- Part 1: Getting Started
- Part 2: Mapping Demographic Information
- Part 3: Mapping Repeating Information
- Part 4: Putting a Channel into Production
- Part 5: Mapping Weight
- Part 6: Filtering Messages (no video)
- Part 7: Processing Lab Messages (no video)
Introduction
These videos for the Translator quickly introduce some core concepts. They are a good place to start when you are learning the Translator and want to get started quickly. After you have gone through them though it is worth going through the rest of the tutorials like the HL7 to Database Tutorial, the Database to HL7 Tutorial and go through resources on Lua which explain the details in much more depth.
Note: These videos were made using Iguana 5.0.2, so the look and feel has changed, and the latest features are not available.
However this is still a very useful resource as the core concepts presented remain unchanged.
Initial Video – Covering the core concepts of the Iguana Translator
This video highlights four core features of the Iguana Translator; Auto-completion, Annotations, Sample Data and Source Control. Learn about the thought process behind this new platform and discover how these features work together to create an innovative new integration platform.
- Introducing the Translator – Initial Introduction.
Tutorial Series #1 – Everything you need to know to map HL7 into a database.
In our first Iguana 5 tutorial series we walk you step-by-step through the process of creating a realistic inbound interface; specifically, an interface that can receive a stream of HL7 messages and process the information into a database. We start by showing you how to setup your environment to follow along and then we move from simple tasks like mapping demographics onto more complex integration tasks like mapping repeating fields.
Part 1: Getting Started [top]
Note: The code in this tutorial uses a legacy db.merge call.
If you are using Iguana 5.5 it is recommended best practice to use database connection objects.
The code on this page will work with all versions of Iguana including 5.5 – it just uses older style functions.
The first in a series of Iguana 5 tutorial videos. In this video, you will be introduced to the Iguana Translator and learn how to setup your environment so that you can follow along and create the same interface on your own machine.Runtime: 7:10
To follow along with our series of videos demonstrating how to build a realistic inbound interface – capable of receiving a steam of HL7 messages and parsing out patient demographic and lab information – you’ll need to setup your environment to match ours.
Luckily, it’s pretty easy. To get started, you’re going to need:
- A copy of Iguana 5
- Access to a local database. We’re using MySQL, and you can download a copy of the Community Server and the Workbench (GUI tool) for free. Or, just use whatever database you already have setup.
- The two files below: Sample.txt and Tutorial.vmd.
Sample Data: sample_data.txt
VMD: tutorial.vmd
Once you have the all the required items, watch the video, follow the steps and then move on to the next video in the series.
Part 2: Mapping Demographic Information [top]
The second in a series of Iguana 5 tutorial videos. In this video, we begin the process of creating our interface. We start by looking at the incoming HL7 message and our database tables. Then, we move on to parsing our message and saving some basic demographic fields.
Runtime: 14:48
In our first video, we showed you how to setup your environment to follow along with the tutorials. So in this video, we begin the process of creating our interface. You can start by coping the script below and pasting it into the Iguana Translator channel you created in Part 1.
Once you have that in place, you can play this video and follow along as we use the Iguana Translator for the first time to parse our HL7 message into a logic data structure, learn how we use our VMD file to create a local table structure and then move on to organizing our code and mapping some basic patient demographic information.
When we’re done, we’ll have a working, though limited, interface that is capable of receiving ADT (patient demographics) messages and saving some of the information into our database.
Note: In this video I was using a required module called “hl7date”. With the official release of Iguana 5, we have replaced hl7date with a much more robust dateparse. The code below is identical to what I was using in my video, but I’ve updated it to use the dateparse module.
Script:
--require("hl7date")
require("dateparse")
-- The main function is the first function called from Iguana.
-- The Data argument will contain the message to be processed.
function main(Data)
--local T = MapData(Data)
--if T then
-- db.merge{api=db.MY_SQL,name='test', user='user', password='pass', data=T}
--end
end
function MapData(Data)
--local Msg,Name = hl7.parse({vmd='tutorial.vmd', data=Data})
--local Tables = db.tables({vmd='tutorial.vmd', name=Name})
--if Name == 'ADT' then
-- ProcessADT(Tables, Msg)
-- return Tables
--end
--return nil
end
-- #### Common Routines ####
function MapPatient(Table, PID)
end
-- #### End of Common Routines ####
-- ##### Processing ADT (Admit/Discharge/Transfer) #####
function ProcessADT(Tables, Msg)
end
-- ##### End of ADT Section #####
Part 3: Mapping Repeating Information [top]
The third in a series of Iguana 5 tutorial videos. In this video, we build on our previous example and begin looking at more complex mapping; specifically repeating next of kin information. We learn how to create our first “loop”, how to use the print statement and annotations to understand our data and end with a second set of information mapped to our database.
Runtime: 11:31
We continue building on the interface we created in our previous video – Mapping Demographic Information – and begin adding the functionality to process repeating, next of kin information.
Throughout the video we:
- Use print statements to show the number of relatives – # count operator and string concatenation .. operator
Note: We now recommend using trace() instead of print(). - Explain how repetition is always just dealing with for loops
- Show navigation controls on Next of Kin function – 1 of N
- Look at the foreign key – patient ID
As an added point, this screen snap is good at explaining the local variable concept. Notice how “i” is nil outside of the scope of the for loop.
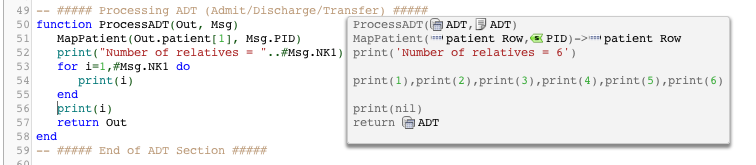
One thing to keep in mind if you plan to use for loops for repeating segments and repeating groups of segments in Iguana is to get the segment grammar correct as defined in the vmd file. For instance if you had a lab result with a repeating set of OBX segments but in your message grammar defined in the vmd file this was defined as a non repeating OBX segment, then you would only see the first OBX segment in the code. To fix that it would be necessary to edit the vmd file in Chameleon and alter the defined grammar to mark the segment as repeating.
Part 4: Putting a Channel into Production [top]
The fourth in a series of Iguana 5 tutorial videos. In this video, we take the interface we created in our previous video and put it “into production”. That means we turn the channel on and send some messages through to see how they are process, logged and saved in the database.
Runtime: 11:31
In this video, we take the interface we were working on in our previous video and put it into production.
Some of the topics covered in this tutorial include:
- The choice of milestone; the option of rigorous change control vs. more relaxed policy when deploying interfaces,
- how to use the iNTERFACEWARE HL7 Simulator to feed in data into your interface,
- how we see the related log entries,
- how to resubmit message, and
- how to turn a message in the logs into sample data for the Iguana Translator.
When we’re done, we’ve successfully processed over 200 messages and automatically created records in our database for both the patient and next of kin information.
Part 5: Mapping Weight
The fifth in a series of Iguana 5 tutorial videos. In this video, we continue our interface design by adding the ability to find and map a patient’s weight. Though seemingly simple, this is actually an example of a non-trivial mapping.
Runtime: 12:59
This tutorial is about showing the power of what a scripting based mapper can do. While we’re using a specific example from HL7 with patient weight mapping, the same concepts can be applied to many different problems.
Patient weight and height are not actually properties of the PID patient identification segment like you might think. They are considered lab results in HL7. So the appear in their own OBX segment.
- Example of a non trivial mapping
- Introduce the problem – show the data.
- Introduce concept of the engine libraries being very minimal
- Put in require function for hl7util
- Show editing of the ObxWeightFilter function
- Explain about the :S() casting to a string – we have a “Node Tree” which is not a string so to compare it we have to convert it a string
This is very specific example of very generic kind of problem that one faces in HL7 often. For instance dealing with babies and looping through the next of kin to find:
- The Mother
- The legal guardian – not always the same as the mother in case of mental illness or drug addiction.
Another good example of this type of problem is phone numbers coming a modern system like an iPhone vs. an older system which just had home phone and business phone.
References:
Part 6: Filtering Messages (no video) [top]
Note: The code on this page uses a legacy db.merge call.
If you are using Iguana 5.5 it is recommended best practice to use database connection objects.
The code on this page will work with all versions of Iguana including 5.5, it just uses older style functions.
Filtering is a very common activity performed by interface engineers. Hospitals often don’t have the ability to easily filter data on their end and so one often has to filter out unwanted messages, or messages relating to facilities that we don’t want to cover.
Using Iguana as an interface makes it very easy to perform this type of filtering, before the data is used by hospital systems. It is very easy to follow best practises, like keeping all the filter code in one place, logging clear informational error messages, structuring code in a simple way to make it clearer for non-technical users, etc.
- All filtering is done in the filterMessage() function. If more complex filtering is needed filterMessage() could call functions like filterMsgRule1(), filterMsgRule2() etc.
- Logging user friendly error messages. In this code the ReportFilter() function is used to send messages to the Iguana log.
- Use of a helper utility deliberately placed near the top of the code can be very helpful since it creates a custom GUI. ReportFilter function is deliberately placed at top, makes it very visible to an non technical analyst as to why a message was filtered.
require("dateparse")
require("hl7util")
require("stringutil")
-- The main function is the first function called from Iguana.
-- The Data argument will contain the message to be processed.
function main(Data)
local T = MapData(Data)
if T then
db.merge{api=db.MY_SQL,name='test',
user='root', password='pass', data=T}
end
end
function ReportFilter(Reason)
print("Message filtered: "..Reason)
end
function MapData(Data)
local Msg,Name = hl7.parse{vmd='demo.vmd', data=Data}
local Out = db.tables{vmd='demo.vmd', name=Name}
if FilterMessage(Msg) then return end
if Name == 'ADT' then
ProcessADT(Out, Msg)
return Out
end
return nil
end
function FilterMessage(Msg)
if Msg:nodeName() == 'Catchall' then
ReportFilter("Unknown message type: "..Msg.MSH[9]:S())
return true
end
if Msg.PID[5][1][1][1]:nodeValue() == 'Muir' then
ReportFilter("Muir family is not welcome here.")
return true
end
return false
end
-- #### Common Routines ####
function MapPatient(T, PID)
T.Id = PID[3][1][1]
T.LastName = PID[5][1][1][1]:nodeValue():capitalize()
T.GivenName = PID[5][1][2]
T.Dob = PID[7][1]:D()
T.Sex = PID[8]
T.Race = PID[10][1][1]
T.Ssn = PID[19]
return T
end
-- #### End of Common Routines ####
function MapKin(T, NK1, Id)
T.PatientId = Id
T.LastName = NK1[2][1][1][1]
T.FirstName = NK1[2][1][2]
T.Relationship = NK1[3][1]
return T
end
-- ##### Processing ADT (Admit/Discharge/Transfer) #####
function ProcessADT(Out, Msg)
MapPatient(Out.patient[1], Msg.PID)
print("Number of relatives = "..#Msg.NK1)
for i=1,#Msg.NK1 do
MapKin(Out.kin[i], Msg.NK1[i], Msg.PID[3][1][1])
end
MapWeight(Out.patient[1], Msg)
return Out
end
function MapWeight(T, Msg)
local OBX = hl7util.findSegment(Msg, FilterObxWeight)
T.Weight = OBX[5][1][1]
end
function FilterObxWeight(S)
if S:nodeName() =='OBX' then
if S[3][1]:nodeValue() == "WT" then
return true
end
end
end
-- ##### End of ADT Section #####
Note: The ReportFilter() function uses print() to log messages. When an Iguana channel is run print() messages are logged as informational messages. For more sophisticated logging you can use the iguana.logInfo(), iguana.logWarning(), iguana.logDebug() functions.
Part 7: Processing Lab Messages (no video) [top]
So let’s look at some general concepts that come up. How to reuse functions, how to generalize code. These just follow best practice for good program structure.
- The lab section of this code re-uses the
MapPatient()function - We also show how
hl7util.findSegment()
Note: this lab.vmd already has a lab message defined (or you can add a lab message to the demo.vmd with Chameleon)
require("hl7date")
require("hl7util")
require("stringutil")
-- The main function is the first function called from Iguana.
-- The Data argument will contain the message to be processed.
function main(Data)
local T = MapData(Data)
if T then
db.merge{api=db.MY_SQL,name='test',
user='root', password='pass', data=T}
end
end
function ReportFilter(Reason)
print("Message filtered: "..Reason)
end
-- Utility node methods to convert HL7 dates (D) and timestamps (T) to database date/time format.
-- Best practice is to define them locally so that they can be customized for interfaces with non-standard date/time formats.
function node.D(DateNode)
return hl7date.Hl7DateToDb(DateNode)
end
function node.T(TimeNode)
return hl7date.Hl7TimeStampToDb(TimeNode)
end
function MapData(Data)
local Msg,Name = hl7.parse{vmd='lab.vmd', data=Data}
local Out = db.tables{vmd='lab.vmd', name=Name}
if FilterMessage(Msg) then return end
if Name == 'ADT' then ProcessADT(Out, Msg)
elseif Name == 'Lab' then ProcessLab(Out,Msg) end
return nil
end
function FilterMessage(Msg)
if Msg:nodeName() == 'Catchall' then
ReportFilter("Unknown message type: "..Msg.MSH[9]:S())
return true
end
if Msg:nodeName() == 'ADT' and Msg.PID[5][1][1][1]:nodeValue() == 'Muir' then
ReportFilter("Muir family is not welcome here.")
return true
end
return false
end
-- #### Common Routines ####
function MapPatient(T, PID)
T.Id = PID[3][1][1]
T.LastName = PID[5][1][1][1]:nodeValue():capitalize()
T.GivenName = PID[5][1][2]
T.Dob = PID[7][1]:D()
T.Sex = PID[8]
if #PID[10] > 0 then
T.Race = PID[10][1][1]
end
T.Ssn = PID[19]
return T
end
-- #### End of Common Routines ####
function MapKin(T, NK1, Id)
T.PatientId = Id
T.LastName = NK1[2][1][1][1]
T.FirstName = NK1[2][1][2]
T.Relationship = NK1[3][1]
return T
end
-- ##### Processing ADT (Admit/Discharge/Transfer) #####
function ProcessADT(Out, Msg)
MapPatient(Out.patient[1], Msg.PID)
print("Number of relatives = "..#Msg.NK1)
for i=1,#Msg.NK1 do
MapKin(Out.kin[i], Msg.NK1[i], Msg.PID[3][1][1])
end
MapWeight(Out.patient[1], Msg)
return Out
end
function MapWeight(T, Msg)
local OBX = hl7util.findSegment(Msg, FilterObxWeight)
T.Weight = OBX[5][1][1]
end
function FilterObxWeight(S)
if S:nodeName() =='OBX' then
if S[3][1]:nodeValue() == "WT" then
return true
end
end
end
-- ##### End of ADT Section #####
-- ## Lab Section
function ProcessLab(Out, Msg)
MapPatient(Out.patient[1], Msg.PATIENT.PID)
-- A second way of doing it.
local PID = hl7util.findSegment(Msg, FilterPID)
MapPatient(Out.patient[1], PID)
MapOrder(Out.order[1], Msg.ORDER[1].ORDER_DETAIL.OBR)
MapResults(Out.result, Msg.ORDER[1].ORDER_DETAIL.OBSERVATION,
Msg.ORDER[1].ORDER_DETAIL.OBR[3][1])
return Out
end
function FilterPID(S)
if S:nodeName() =='PID' then
return true
end
end
function MapOrder(T, OBR)
T.OrderFillerNumber = OBR[3][1]
return T
end
function MapResults(T, R, OrderNum)
for i=1, #R do
MapResult(T[i], R[i], OrderNum)
end
end
function MapResult(T, R, OrderNum)
T.OrderFillerNumber = OrderNum
-- TODO map some OBX fields.
local N =''
for i=1, #R.NTE do
N = N..R.NTE[i]:S()
end
T.Notes = N
end
What’s next?
Need a better lab result message example which has NTEs associated with lab results.
Need to flesh out the lab.vmd?
Here’s a nice realistic lab message contributed by a customer. Confidential patient information has been removed:
MSH|^~\&|FDHL7|JOHNSON LABS||P1055|201007231634||ORU^R01|P1055-0000047907|P|2.3|1||NE|NE PID|1|JQ4988|108512373||SAMPLES^JUNIOR||01/10/1948^53 Y|M|||^******^^|||||||| NTE|1|P|**************************************************************************** ADD|NON FASTING OBR|1||108512373|CHEM^-------* CHEMISTRY *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0135-4^Total Protein||7.3|gm/dl|5.9-8.4||||F OBX|2|NM|0033-1^Albumin||3.9|gm/dl|3.2-5.2||||F OBX|3|NM|1753-3^Globulin||3.4|gm/dL|1.7-3.7||||F OBX|4|NM|0641-1^A/G Ratio||1.1||1.1-2.9||||F OBX|5|NM|1976-0^Glucose||296|mg/dL|70-99|HI|||F OBX|6|NM|0148-7^Sodium||134|mmol/L|133-145||||F OBX|7|NM|0129-7^Potassium||4.3|mmol/L|3.3-5.3||||F OBX|8|NM|0057-0^Chloride||96|mmol/L|96-108||||F OBX|9|NM|0052-1^CO2||24|mmol/L|21-29||||F OBX|10|NM|0049-7^BUN||17|mg/dl|7-25||||F OBX|11|NM|0070-3^Creatinine||1.1|mg/dl|0.6-1.3||||F OBX|12|NM|090013-4^e-GFR||70||> 60 mL/min/1.73m2||||F OBX|13|NM|1427-4^BUN/Creat Ratio||15.5||10-28||||F OBX|14|NM|0050-5^Calcium||8.9|mg/dl|8.4-10.4||||F OBX|15|NM|0157-8^Uric Acid||6.2|mg/dl|2.4-7.0||||F OBX|16|NM|0114-9^Iron||87|mcg/dl|30-160||||F OBX|17|NM|0043-0^Bilirubin, Total||0.6|mg/dl|0.1-1.0||||F OBX|18|NM|0117-2^LDH||190|u/l|94-250||||F OBX|19|NM|0185-9^Alk Phos||63|u/l|39-120||||F OBX|20|NM|0146-1^AST (SGOT)||33|u/l|0-37||||F OBX|21|NM|0127-1^Phosphorous||2.8|mg/dl|2.6-4.5||||F OBX|22|NM|0147-9^ALT (SGPT)||55|u/L|0-40|HI|||F OBX|23|NM|0093-5^G-GTP||33|u/L|7-51||||F NTE|1|L|**************************************************************************** ADD|GFR (Glomerular Filtration Rate) calculation utilizes the MDRD formula ADD|(Modification of Diet in Renal Disease Study Group) and assumes a normal ADD|adult body surface area of 1.73. If the patient is African American ADD|multiply result reported by 1.21. (Ref. National Kidney Disease Educa. ADD|Program.) ADD| ***** Male/Female reference range: >60 mL/min/1.73 m2 ***** ADD|Note: A calculated GFR of <60 mL suggests chronic kidney disease, but ADD|only if found consistently over at least 3 months. A calculated ADD|result of <15 mL is consistent with renal failure. OBR|2||108512373|CARD^-* CARDIOVASCULAR/LIPIDS *--||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0058-8^Cholesterol||124|mg/dl|< 200||||F OBX|2|NM|0155-2^Triglycerides||73|mg/dl|< 151||||F OBX|3|NM|0059-6^HDL CHOL.,DIRECT||39|mg/dl|>40|LO|||F OBX|4|NM|1764-0^HDL as % of Cholesterol||31|%|||||F NTE|1|L|Range/Evaluation: (> 25) BELOW AVERAGE RISK OBX|5|NM|1421-7^Chol/HDL Ratio||3.18| |||||F NTE|1|L|Range/Evaluation: (<4.2) BELOW AVERAGE RISK OBX|6|NM|0253-5^LDL/HDL Ratio||1.82| |0-3.55||||F OBX|7|NM|0505-8^LDL Cholesterol||71||< 100||||F OBX|8|NM|3345-6^VLDL, CALCULATED||14|mg/dl|7-32||||F OBR|3||108512373|HEMA^------* HEMATOLOGY *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|1497-7^WBC||6.61|x10(3)/uL|3.40-11.80||||F OBX|2|NM|1498-5^RBC||4.56|x10(6)/uL|4.20-5.90||||F OBX|3|NM|1499-3^HGB||13.6|gm/dL|12.3-17.0||||F OBX|4|NM|0019-0^HCT||39.9|%|39.3-52.5||||F OBX|5|NM|1503-2^MCV||87.5|fL|80.0-100.0||||F OBX|6|NM|1504-0^MCH||29.8|pg|25.0-34.1||||F OBX|7|NM|1502-4^MCHC||34.1|gm/dL|29.0-35.0||||F OBX|8|NM|1598-2^RDW||14.1|%|10.9-16.9||||F OBX|9|NM|1505-7^POLYS||58.8|%|36.0-78.0||||F OBX|10|NM|3176-5^POLYS, ABS. COUNT||3.89|x10(3)/uL|1.22-9.20||||F OBX|11|NM|1507-3^LYMPHS||31.0|%|12.0-48.0||||F OBX|12|NM|3177-3^LYMPHS, ABS. COUNT||2.05|x10(3)/uL|0.41-5.66||||F OBX|13|NM|1511-5^MONOS||7.7|%|0.0-13.0||||F OBX|14|NM|3180-7^MONOS, ABS. COUNT||0.51|x10(3)/uL|0.17-1.42||||F OBX|15|NM|1509-9^EOS||2.0|%|0.0-8.0||||F OBX|16|NM|3178-1^EOS, ABS. COUNT||0.13|x10(3)/uL|0.03-0.94||||F OBX|17|NM|1510-7^BASOS||0.3|%|0.0-2.0||||F OBX|18|NM|3179-9^BASOS, ABS. COUNT||0.02|x10(3)/uL|0.00-0.24||||F OBX|19|NM|270053-2^IMMATURE GRANULOCYTES||0.2|%|0.0-0.5||||F OBX|20|NM|0128-9^PLATELET COUNT||191|x10(3)/uL|144-400||||F OBX|21|NM|400053-5^MPV||10.6|fL|8.2-11.9||||F OBR|4||108512373|URIN^------* URINALYSIS *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|ST|6315-6^Color||YELLOW||YELLOW, STRAW, AMBER||||F OBX|2|ST|6316-4^Character||CLEAR||CLEAR||||F OBX|3|NM|1520-6^Specific Gravity URN||1.030||1.003 - 1.030||||F OBX|4|NM|1521-4^pH Urine||5.5||5.0 - 8.0||||F OBX|5|ST|1522-2^Protein, Urine||NEGATIVE||NEGATIVE||||F OBX|6|ST|1523-0^Glucose, Urine||3+,>=1000 mg/dL||NEGATIVE|*|||F OBX|7|ST|1524-8^Ketone, Urine||NEGATIVE||NEGATIVE||||F OBX|8|NM|1525-5^Urobilinogen Urine||1.0|Units|0.2 - 1.0||||F OBX|9|ST|1526-3^Bilirubin, Urine||NEGATIVE||NEGATIVE||||F OBX|10|ST|1527-1^Blood, Urine||NEGATIVE||NEGATIVE||||F OBX|11|ST|1528-9^Nitrites Urine||NEGATIVE||NEGATIVE||||F OBX|12|ST|6311-5^Leukocyte Esterase||NEGATIVE||NEGATIVE||||F OBX|13|ST|1529-7^Crystals Urine||NONE||NONE||||F OBX|14|ST|2135-2^Crystal Amt. Urine||NONE||NONE||||F OBX|15|ST|1534-7^WBC, Urine||0-4|PER HPF|0-4||||F OBX|16|ST|1535-4^RBC, Urine||0-3|PER HPF|0-3||||F OBX|17|ST|1546-1^Epithelial Cells, Ur||FEW||FEW||||F OBX|18|ST|1545-3^Cast, Hyaline, Urine||NONE SEEN|PER LPF|0-4||||F OBX|19|ST|1547-9^Cast, Granular, Urin||NONE SEEN|PER LPF|0-1||||F OBX|20|ST|1543-8^Cast, RBC, Urine||NONE SEEN|PER LPF|0-1||||F OBX|21|ST|1549-5^Bacteria, Urine||NONE||FEW||||F NTE|1|L|**************************************************************************** ADD|NOTE: Significant quantities of epithelial cells will ADD|be identified if they are not squamous cell types. OBR|5||108512373|MISC^-----* MISCELLANEOUS *------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0153-7^TSH||1.930||0.27-4.2 uIU/mL||||F OBX|2|NM|0151-1^THYROXINE(T4)||9.3||4.5-12.0 ug/dL||||F OBX|3|NM|0152-9^T3 UPTAKE||29.7||24.3-39.0%||||F OBX|4|NM|0666-8^FREE T4 INDEX||2.8| |1.1-4.5||||F OBX|5|ST|0142-0^RPR||NON-REACT||NON-REACTIVE||||F NTE|1|L|**************************************************************************** ADD|NOTICE: IF the result of the RPR is reported as reactive with a titer ADD|of up to 1:8 please note that this level of reactivity can be caused ADD|by other, non-specific constituents and may not be related to syphilis. ADD|Confirmation of positive RPRs can only be made via performance of the ADD|T. Pallidum confirmation test. OBX|6|NM|0102-4^HGB. A1c(glycohgb)||9.1||4-6%|HI|||F OBX|7|ST|1661-8^CREAT.URN.TIMED/RAND||.147||gms/dL||||F OBX|8|NM|2699-7^MICROALB/CREAT RATIO||4.1||<30mg/gm creat.||||F OBX|9|NM|3172-4^MICROALBUMIN,RANDOM||0.6||<2.9 mg/dL||||F NTE|2|L|**************************************************************************** ADD|GLYCOHEMOGLOBIN(HgbA1c)Ranges% eAG ranges(mg/dL)* GLUCOSE CONTROL INDEX ADD| ADD| < 4-6% <68-126 Non-Diabetic Level ADD| < 6-7% <126-154 Diabetic Control ADD| > 8% >183 Additional action suggested ADD|*Data adapted from the A1c-Derived Average Glucose (ADAG) Study ADD|(2006-2008). Estimated average glucose (eAG) values (shown as ranges ADD|in the above table) can be reported as individual patient values if ADD|requested. NTE|3|L|**************************************************************************** ADD|NOTE: SST tube submitted was inadequately spun. Serum was found to ADD|contain RBCs. Certain tests, e.g. Glucose, may be decreased while ADD|others e.g. Potassium or LDH may be elevated. FTS|1|END OF FILE
A trap for young players who are still learning about HL7 (like me), “Implicit Segments” that are not visible when inspecting message data manually. The “PATIENT” segment in the Lab Section of the code is an example.
HL7 messages have two parts the message definition which is complete and data which is sparse. This means that to understand the structure of an HL7 message you need to look at the message definition, not the data (unlike XML where the data structure is explicit).
As you can see the “implicit segment” is visible in the message definition and the code, but not visible in the data.
You can also see this by looking at the VMD Message Grammar in Chameleon:



