- Introduction
- In theory you should not need to convert date/time values, but in practice…
- A real world lab message with non-standard date/time format
- Non-conformant Lab Result Message
- What the bare bones of the Translator gives you
- Using the fuzzy date/time parser
- Ambiguous Date/Time Formats
- Complete documentation for the fuzzy date/time parser
- Using os.date to output arbitrary date/time formats
- Don’t be afraid to mix in a bit of string parsing
- Some user suggestions for other useful date/time libraries
Introduction
Because this is an older article it was written for Iguana 5 so it uses Iguana 5 screenshots, and may contain some outdated references.
The core of the Iguana Translator has very little built in support for date and time formats. Every node is treated as a string. If you try to assign a date/time of the wrong format to a date/time field in a table an error will be raised.
This is deliberate part of the minimalist design philosophy of the Translator.
However one of our rockstars has written a fuzzy date/time parser in Lua which parses just about any date. It’s very nice since it means except for a few special cases you can avoid the need to ever have to explicitly worry about converting date/time formats. Less than 300 lines of simple code that will save literally millions of dollars globally in the industry on unnecessary busy work.
You can download the latest code from the Builtin: Iguana Date/Time repository, see this article: Parse any date time value.
If you have any questions please contact us at support@interfaceware.com.
Download dateparse.lua from our code repository
The source code for the fuzzy date/time parser can be downloaded from the Date/Time repository. To use it you just need to import the channel.
In theory you should not need to convert date/time values, but in practice… [top]
HL7 does actually clearly state that dates and times should by formatted in this format:
yyyy[mm[dd[HHMM[SS[.ssss]]]]][zzzz]
So if all implementations followed the standard, integration engineers shouldn’t have to deal with converting incompatible date time formats.
But in practice real implementations of HL7 do break the standard like this lab message. Also there are older pre-HL7 formats that are even worse in terms of containing a mix of formats in the same message.
So it this is a problem that has to dealt with. Fortunately an elegant solution exists.
A real world lab message with non-standard date/time format [top]
An example of real world lab message. Notice the non standard date/time format?
MSH|^~\&|FDHL7|JOHNSON LABS||P1055|201007231634||ORU^R01|P1055-0000047907|P|2.3|1||NE|NE PID|1|JQ4988|108512373||SAMPLES^JUNIOR||01/10/1948^53 Y|M|||^******^^|||||||| NTE|1|P|**************************************************************************** ADD|NON FASTING OBR|1||108512373|CHEM^-------* CHEMISTRY *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0135-4^Total Protein||7.3|gm/dl|5.9-8.4||||F OBX|2|NM|0033-1^Albumin||3.9|gm/dl|3.2-5.2||||F OBX|3|NM|1753-3^Globulin||3.4|gm/dL|1.7-3.7||||F OBX|4|NM|0641-1^A/G Ratio||1.1||1.1-2.9||||F OBX|5|NM|1976-0^Glucose||296|mg/dL|70-99|HI|||F OBX|6|NM|0148-7^Sodium||134|mmol/L|133-145||||F OBX|7|NM|0129-7^Potassium||4.3|mmol/L|3.3-5.3||||F OBX|8|NM|0057-0^Chloride||96|mmol/L|96-108||||F OBX|9|NM|0052-1^CO2||24|mmol/L|21-29||||F OBX|10|NM|0049-7^BUN||17|mg/dl|7-25||||F OBX|11|NM|0070-3^Creatinine||1.1|mg/dl|0.6-1.3||||F OBX|12|NM|090013-4^e-GFR||70||> 60 mL/min/1.73m2||||F OBX|13|NM|1427-4^BUN/Creat Ratio||15.5||10-28||||F OBX|14|NM|0050-5^Calcium||8.9|mg/dl|8.4-10.4||||F OBX|15|NM|0157-8^Uric Acid||6.2|mg/dl|2.4-7.0||||F OBX|16|NM|0114-9^Iron||87|mcg/dl|30-160||||F OBX|17|NM|0043-0^Bilirubin, Total||0.6|mg/dl|0.1-1.0||||F OBX|18|NM|0117-2^LDH||190|u/l|94-250||||F OBX|19|NM|0185-9^Alk Phos||63|u/l|39-120||||F OBX|20|NM|0146-1^AST (SGOT)||33|u/l|0-37||||F OBX|21|NM|0127-1^Phosphorous||2.8|mg/dl|2.6-4.5||||F OBX|22|NM|0147-9^ALT (SGPT)||55|u/L|0-40|HI|||F OBX|23|NM|0093-5^G-GTP||33|u/L|7-51||||F NTE|1|L|**************************************************************************** ADD|GFR (Glomerular Filtration Rate) calculation utilizes the MDRD formula ADD|(Modification of Diet in Renal Disease Study Group) and assumes a normal ADD|adult body surface area of 1.73. If the patient is African American ADD|multiply result reported by 1.21. (Ref. National Kidney Disease Educa. ADD|Program.) ADD| ***** Male/Female reference range: >60 mL/min/1.73 m2 ***** ADD|Note: A calculated GFR of <60 mL suggests chronic kidney disease, but ADD|only if found consistently over at least 3 months. A calculated ADD|result of <15 mL is consistent with renal failure. OBR|2||108512373|CARD^-* CARDIOVASCULAR/LIPIDS *--||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0058-8^Cholesterol||124|mg/dl|< 200||||F OBX|2|NM|0155-2^Triglycerides||73|mg/dl|< 151||||F OBX|3|NM|0059-6^HDL CHOL.,DIRECT||39|mg/dl|>40|LO|||F OBX|4|NM|1764-0^HDL as % of Cholesterol||31|%|||||F NTE|1|L|Range/Evaluation: (> 25) BELOW AVERAGE RISK OBX|5|NM|1421-7^Chol/HDL Ratio||3.18| |||||F NTE|1|L|Range/Evaluation: (<4.2) BELOW AVERAGE RISK OBX|6|NM|0253-5^LDL/HDL Ratio||1.82| |0-3.55||||F OBX|7|NM|0505-8^LDL Cholesterol||71||< 100||||F OBX|8|NM|3345-6^VLDL, CALCULATED||14|mg/dl|7-32||||F OBR|3||108512373|HEMA^------* HEMATOLOGY *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|1497-7^WBC||6.61|x10(3)/uL|3.40-11.80||||F OBX|2|NM|1498-5^RBC||4.56|x10(6)/uL|4.20-5.90||||F OBX|3|NM|1499-3^HGB||13.6|gm/dL|12.3-17.0||||F OBX|4|NM|0019-0^HCT||39.9|%|39.3-52.5||||F OBX|5|NM|1503-2^MCV||87.5|fL|80.0-100.0||||F OBX|6|NM|1504-0^MCH||29.8|pg|25.0-34.1||||F OBX|7|NM|1502-4^MCHC||34.1|gm/dL|29.0-35.0||||F OBX|8|NM|1598-2^RDW||14.1|%|10.9-16.9||||F OBX|9|NM|1505-7^POLYS||58.8|%|36.0-78.0||||F OBX|10|NM|3176-5^POLYS, ABS. COUNT||3.89|x10(3)/uL|1.22-9.20||||F OBX|11|NM|1507-3^LYMPHS||31.0|%|12.0-48.0||||F OBX|12|NM|3177-3^LYMPHS, ABS. COUNT||2.05|x10(3)/uL|0.41-5.66||||F OBX|13|NM|1511-5^MONOS||7.7|%|0.0-13.0||||F OBX|14|NM|3180-7^MONOS, ABS. COUNT||0.51|x10(3)/uL|0.17-1.42||||F OBX|15|NM|1509-9^EOS||2.0|%|0.0-8.0||||F OBX|16|NM|3178-1^EOS, ABS. COUNT||0.13|x10(3)/uL|0.03-0.94||||F OBX|17|NM|1510-7^BASOS||0.3|%|0.0-2.0||||F OBX|18|NM|3179-9^BASOS, ABS. COUNT||0.02|x10(3)/uL|0.00-0.24||||F OBX|19|NM|270053-2^IMMATURE GRANULOCYTES||0.2|%|0.0-0.5||||F OBX|20|NM|0128-9^PLATELET COUNT||191|x10(3)/uL|144-400||||F OBX|21|NM|400053-5^MPV||10.6|fL|8.2-11.9||||F OBR|4||108512373|URIN^------* URINALYSIS *--------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|ST|6315-6^Color||YELLOW||YELLOW, STRAW, AMBER||||F OBX|2|ST|6316-4^Character||CLEAR||CLEAR||||F OBX|3|NM|1520-6^Specific Gravity URN||1.030||1.003 - 1.030||||F OBX|4|NM|1521-4^pH Urine||5.5||5.0 - 8.0||||F OBX|5|ST|1522-2^Protein, Urine||NEGATIVE||NEGATIVE||||F OBX|6|ST|1523-0^Glucose, Urine||3+,>=1000 mg/dL||NEGATIVE|*|||F OBX|7|ST|1524-8^Ketone, Urine||NEGATIVE||NEGATIVE||||F OBX|8|NM|1525-5^Urobilinogen Urine||1.0|Units|0.2 - 1.0||||F OBX|9|ST|1526-3^Bilirubin, Urine||NEGATIVE||NEGATIVE||||F OBX|10|ST|1527-1^Blood, Urine||NEGATIVE||NEGATIVE||||F OBX|11|ST|1528-9^Nitrites Urine||NEGATIVE||NEGATIVE||||F OBX|12|ST|6311-5^Leukocyte Esterase||NEGATIVE||NEGATIVE||||F OBX|13|ST|1529-7^Crystals Urine||NONE||NONE||||F OBX|14|ST|2135-2^Crystal Amt. Urine||NONE||NONE||||F OBX|15|ST|1534-7^WBC, Urine||0-4|PER HPF|0-4||||F OBX|16|ST|1535-4^RBC, Urine||0-3|PER HPF|0-3||||F OBX|17|ST|1546-1^Epithelial Cells, Ur||FEW||FEW||||F OBX|18|ST|1545-3^Cast, Hyaline, Urine||NONE SEEN|PER LPF|0-4||||F OBX|19|ST|1547-9^Cast, Granular, Urin||NONE SEEN|PER LPF|0-1||||F OBX|20|ST|1543-8^Cast, RBC, Urine||NONE SEEN|PER LPF|0-1||||F OBX|21|ST|1549-5^Bacteria, Urine||NONE||FEW||||F NTE|1|L|**************************************************************************** ADD|NOTE: Significant quantities of epithelial cells will ADD|be identified if they are not squamous cell types. OBR|5||108512373|MISC^-----* MISCELLANEOUS *------||201007221041||||||||201007222312||P1055^SCI DULUTH/PHS^RTE 29,PO BOX 244^DULUTH, MN 19426^|(945)443-1234|RECEPTION, NEW||||201007231634|||R|||| OBX|1|NM|0153-7^TSH||1.930||0.27-4.2 uIU/mL||||F OBX|2|NM|0151-1^THYROXINE(T4)||9.3||4.5-12.0 ug/dL||||F OBX|3|NM|0152-9^T3 UPTAKE||29.7||24.3-39.0%||||F OBX|4|NM|0666-8^FREE T4 INDEX||2.8| |1.1-4.5||||F OBX|5|ST|0142-0^RPR||NON-REACT||NON-REACTIVE||||F NTE|1|L|**************************************************************************** ADD|NOTICE: IF the result of the RPR is reported as reactive with a titer ADD|of up to 1:8 please note that this level of reactivity can be caused ADD|by other, non-specific constituents and may not be related to syphilis. ADD|Confirmation of positive RPRs can only be made via performance of the ADD|T. Pallidum confirmation test. OBX|6|NM|0102-4^HGB. A1c(glycohgb)||9.1||4-6%|HI|||F OBX|7|ST|1661-8^CREAT.URN.TIMED/RAND||.147||gms/dL||||F OBX|8|NM|2699-7^MICROALB/CREAT RATIO||4.1||<30mg/gm creat.||||F OBX|9|NM|3172-4^MICROALBUMIN,RANDOM||0.6||<2.9 mg/dL||||F NTE|2|L|**************************************************************************** ADD|GLYCOHEMOGLOBIN(HgbA1c)Ranges% eAG ranges(mg/dL)* GLUCOSE CONTROL INDEX ADD| ADD| < 4-6% <68-126 Non-Diabetic Level ADD| < 6-7% <126-154 Diabetic Control ADD| > 8% >183 Additional action suggested ADD|*Data adapted from the A1c-Derived Average Glucose (ADAG) Study ADD|(2006-2008). Estimated average glucose (eAG) values (shown as ranges ADD|in the above table) can be reported as individual patient values if ADD|requested. NTE|3|L|**************************************************************************** ADD|NOTE: SST tube submitted was inadequately spun. Serum was found to ADD|contain RBCs. Certain tests, e.g. Glucose, may be decreased while ADD|others e.g. Potassium or LDH may be elevated. FTS|1|END OF FILE
Non-conformant Lab Result Message [top]
I can’t say where this comes from. Save to say that it’s from a big laboratory vendor.
This is nothing unusual in healthcare. The least conformant interfaces are the ones from the biggest most dominant vendors in the industry. Often they are stuck on ancient systems that were written long before even HL7 existed.
Notice the mix of date time formats. The date formats are somewhat ambiguous. See the DD-MMM-YY date format? Not terribly good for the turn of century. Ah well this is the reality of the systems we have to interface with in healthcare. I’ve attached a vmd and some Lua code that works with this.
I used the fuzzy date/time parser.
A^70^20090402^06:51^00012^00280^00150^ B^PD81020^70CM210317^20090121^RACHELLE^SMITH^F^^19830106^F^1322740^DR. M. F. WEATHERHEAD,^2494 DANFORTH AVE^TORONTO,ONT, ONTARIO^M4C 1K9^^1475^^^416-726-4505^416-698-3898^21-JAN-09^ C^CYTOLOGY^^^CLINICAL DATA^A^^^^^70^Y^ D^CYTOLOGY^^ ^ ^ ^ C^CYTOLOGY^^^DATE OF L.M.P.^A^^^^NONE GIVEN^70^N^ C^CYTOLOGY^^^^^^^^^^Y^ D^CYTOLOGY^^ NO OTHER DATA GIVEN^70^ ^ D^CYTOLOGY^^ ^ ^ ^ D^CYTOLOGY^^ ****************************************^ ^ ^ D^CYTOLOGY^^ ^ ^ ^ C^CYTOLOGY^^^PAP SMEAR^N^^^^^70^Y^ D^CYTOLOGY^^ SATISFACTORY FOR EVALUATION.^ ^ ^ D^CYTOLOGY^^ TRANSFORMATION ZONE COMPONENT: ABSENT^ ^ ^ D^CYTOLOGY^^ NEGATIVE FOR INTRAEPITHELIAL LESION OR ^ ^ ^ D^CYTOLOGY^^ MALIGNANCY.^ ^ ^ D^CYTOLOGY^^ SHIFT IN FLORA CONSISTENT WITH BACTERIAL^ ^ ^ D^CYTOLOGY^^ VAGINOSIS.^ ^ ^ D^CYTOLOGY^^ ^ ^ ^ C^CYTOLOGY^^^^^^^^^^Y^ D^CYTOLOGY^^ ^70^ ^ D^CYTOLOGY^^ ****************************************^ ^ ^ D^CYTOLOGY^^ ^ ^ ^ C^CYTOLOGY^^^CYTOTECHNOLOGIST^N^^^^FH^70^Y^ D^CYTOLOGY^^ ^ ^ ^ B^PD84151^^20090401^DAVID^CHUA^M^6586893619LL^19331001^P^1322740^DR. M. F. WEATHERHEAD,^2494 DANFORTH AVE^TORONTO,ONT, ONTARIO^M4C 1K9^^1475^^^416-449-1916^416-698-3898^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^A^135^180^G/L^82^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^A^0.37^0.54^L/L^0.279^70^N^ C^HEMATOLOGY^^^WBC COUNT^A^4.0^11.0^X10 9/L^12.3^70^N^ C^HEMATOLOGY^^^RBC COUNT^A^4.50^6.50^X10 12/L^4.33^70^N^ C^HEMATOLOGY^^^MCV^A^80^97^FL^64.5^70^N^ C^HEMATOLOGY^^^MCH^A^27.0^32.0^PG^18.9^70^N^ C^HEMATOLOGY^^^MCHC^A^320^360^G/L^293^70^N^ C^HEMATOLOGY^^^RDW^A^11.0^14.5^%^20.9^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^311^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^A^2.0^7.5^X10 9/L^8.1^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.7^70^N^ C^HEMATOLOGY^^^(A) MONO^A^0.0^0.8^X10 9/L^1.2^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.2^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.1^70^N^ C^HEMATOLOGY^^^HYPOCHROMIA^A^^^^1+^70^N^ C^HEMATOLOGY^^^MICROCYTOSIS^A^^^^3+^70^N^ C^HEMATOLOGY^^^OVALOCYTES^A^^^^1+^70^N^ C^HEMATOLOGY^^^POLYCHROMASIA^A^^^^SL INCREASED^70^N^ C^HEMATOLOGY^^^TARGET CELLS^N^^^^FEW^70^N^ C^HEMATOLOGY^^^PLATELETS^N^^^^NORMAL^70^N^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^A^3.3^6.0^MMOL/L^6.6^70^N^ C^CHEMISTRY^^^CREATININE^N^60^127^UMOL/L^82^70^N^ C^CHEMISTRY^^^eGFR^N^^^^79^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^141^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^4.1^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^105^70^N^ C^CHEMISTRY^^^CHOLESTEROL^N^TARGET <5.20^TARGET <5.20^MMOL/L^3.40^70^N^ C^CHEMISTRY^^^TRIGLYCERIDES^N^TARGET <1.71^TARGET <1.71^MMOL/L^0.87^70^N^ C^CHEMISTRY^^^HDL CHOLESTEROL^A^TARGET >1.29^TARGET >1.29^MMOL/L^0.85^70^N^ C^CHEMISTRY^^^LDL CHOLESTEROL^N^See Targets^See Targets^MMOL/L^2.15^70^N^ C^CHEMISTRY^^^CHOL/HDL RATIO^A^See Targets^See Targets^^4.00^70^Y^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ Clinical Risk Status: Target Lipid Levels^ ^ ^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ If Risk : LDL-C --Targets-- CHOL/HDL Ratio^ ^ ^ D^CHEMISTRY^^ Low : <5.1 and <6.0^ ^ ^ D^CHEMISTRY^^ Moderate : <3.5 and <5.0^ ^ ^ D^CHEMISTRY^^ High : <2.0 and <4.0^ ^ ^ D^CHEMISTRY^^ ...as per 2006 Canadian Guidelines^ ^ ^ C^URINALYSIS^ROUTINE:^^APPEARANCE^N^^^^CLOUDY^70^N^ C^URINALYSIS^ROUTINE:^^COLOUR^N^^^^YELLOW^70^N^ C^URINALYSIS^ROUTINE:^^PH^N^5.0^9.0^^6.0^70^N^ C^URINALYSIS^ROUTINE:^^PROTEIN^N^NEGATIVE^NEGATIVE^G/L^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^GLUCOSE^N^NEGATIVE^NEGATIVE^MMOL/L^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^KETONE^A^NEGATIVE^NEGATIVE^MMOL/L^1.5^70^N^ C^URINALYSIS^ROUTINE:^^BLOOD^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^NITRITE^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^LEUKOCYTE ESTERASE^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^SPECIFIC GRAVITY^N^^^^1.024^70^N^ B^PD84155^^20090401^SANDRA^BLOG^F^2331943635NB^19770119^F^1322740^DR. M. F. WEATHERHEAD,^2494 DANFORTH AVE^TORONTO,ONT, ONTARIO^M4C 1K9^^1475^^^416-694-9364^416-698-3898^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^143^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.422^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^8.5^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.52^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^93.3^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^31.7^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^340^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^14.0^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^347^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^6.2^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^1.9^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.3^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.1^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^CHEMISTRY^^^GLUCOSE RANDOM^N^3.3^7.8^MMOL/L^4.9^70^N^ C^RIA^^^TSH^N^0.30^4.70^MU/L^0.77^70^N^ B^ST34976^^20090401^PAUL^TOSH^M^1940403759VX^19530319^F^1322740^DR. M. F. WEATHERHEAD,^2494 DANFORTH AVE^TORONTO,ONT, ONTARIO^M4C 1K9^^1475^^^416-925-7686^416-698-3898^01-APR-09^ C^HEMATOLOGY^COAGULATION^^I.N.R.^A^0.8^1.2^^2.8^70^Y^ D^HEMATOLOGY^COAGULATION^ ^ ^ ^ D^HEMATOLOGY^COAGULATION^ RECOMMENDED I.N.R. RANGE^ ^ ^ D^HEMATOLOGY^COAGULATION^ ------------------------------------^ ^ ^ D^HEMATOLOGY^COAGULATION^ THROMBOEMBOLISM RISK 2.0 - 3.0^ ^ ^ D^HEMATOLOGY^COAGULATION^ PROSTHETIC HEART VALVE 2.5 - 3.5^ ^ ^ D^HEMATOLOGY^COAGULATION^ ^ ^ ^ B^AT03584^^20090401^JENNIE^MUIRWOOD^F^1976006344RC^19300724^P^2719080^DR. DANIEL H. ARNAUDON^322 SILVERTHORN AVE^TORONTO, ONTARIO^M6N 3K6^^1475^2719080^^416-622-5183^416-656-8777^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^128^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.383^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^9.4^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.16^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^92.2^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^30.7^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^333^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^14.1^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^359^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^4.9^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^3.3^70^N^ C^HEMATOLOGY^^^(A) MONO^A^0.0^0.8^X10 9/L^0.9^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.3^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^CHEMISTRY^^^MAGNESIUM-SERUM^N^0.65^1.05^MMOL/L^0.97^70^N^ C^CHEMISTRY^^^CREATININE^A^60^115^UMOL/L^153^70^N^ C^CHEMISTRY^^^eGFR^A^^^^28^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ eGFR = 15-29 mL/min/1.73 sq.m^ ^ ^ D^CHEMISTRY^^ Consistent with severe chronic kidney disease^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^A^120^400^UMOL/L^516^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^136^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^4.0^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^100^70^N^ C^CHEMISTRY^^^BICARBONATE(CO2)^N^21.0^28.0^MMOL/L^23.0^70^N^ C^CHEMISTRY^^^ALBUMIN^N^33.0^46.0^G/L^39.6^70^N^ C^CHEMISTRY^^^CALCIUM^N^2.15^2.60^MMOL/L^2.33^70^N^ C^CHEMISTRY^^^PHOSPHATE^A^0.90^1.52^MMOL/L^1.57^70^N^ C^RIA^^^TSH^N^0.30^4.70^MU/L^1.11^70^N^ C^RIA^^^FERRITIN^N^13^145^UG/L^25^70^N^ C^^^^^^^^^^^Y^ D^^^Original to DR. A. BERBECE^^ ^ B^AT03599^^20090401^MARY ANNE^KARMA^F^1561061951JF^19530120^P^2719080^DR. DANIEL H. ARNAUDON^322 SILVERTHORN AVE^TORONTO, ONTARIO^M6N 3K6^^1475^^^416-241-0315^416-656-8777^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^127^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.380^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^5.7^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^3.83^70^N^ C^HEMATOLOGY^^^MCV^A^80^97^FL^99.3^70^N^ C^HEMATOLOGY^^^MCH^A^27.0^32.0^PG^33.2^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^335^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^13.6^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^223^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^2.7^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.3^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.5^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.1^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^CHEMISTRY^^^CREATININE^N^60^115^UMOL/L^68^70^N^ C^CHEMISTRY^^^eGFR^N^^^^77^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^N^120^400^UMOL/L^378^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^139^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^3.8^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^101^70^N^ C^URINALYSIS^ROUTINE:^^APPEARANCE^N^^^^CLEAR^70^N^ C^URINALYSIS^ROUTINE:^^COLOUR^N^^^^YELLOW^70^N^ C^URINALYSIS^ROUTINE:^^PH^N^5.0^9.0^^6.0^70^N^ C^URINALYSIS^ROUTINE:^^PROTEIN^N^NEGATIVE^NEGATIVE^G/L^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^GLUCOSE^N^NEGATIVE^NEGATIVE^MMOL/L^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^KETONE^N^NEGATIVE^NEGATIVE^MMOL/L^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^BLOOD^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^NITRITE^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^LEUKOCYTE ESTERASE^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^URINALYSIS^ROUTINE:^^SPECIFIC GRAVITY^N^^^^1.005^70^N^ B^SX69857^^20090401^JOY^COW^F^4442522555BP^19590805^F^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^416-285-0903^905-771-7771^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^122^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^A^0.37^0.47^L/L^0.369^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^5.4^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.06^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^90.9^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^30.0^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^331^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^14.3^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^266^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^2.0^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.8^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.4^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.1^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^N^3.3^6.0^MMOL/L^5.3^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^4.1^70^N^ B^XK64610^^20090328^GEORGIA^MULRANEY^F^9770677731^19800710^F^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^416-859-8978^905-771-7771^28-MAR-09^ C^RIA^^^TSH^N^0.30^4.70^MU/L^2.42^70^N^ C^RIA^^^T4 FREE^N^9.1^23.8^PMOL/L^15.3^70^N^ C^RIA^^^FREE T3^N^2.5^5.7^PMOL/L^3.7^70^N^ C^RIA^^^CORTISOL AM-SERUM^N^101^536^NMOL/L^349^70^N^ C^RIA^^^FREE TESTOSTERONE^N^< 9.0^< 9.0^PMOL/L^4.3^70^N^ C^RIA^^^ESTRADIOL^N^^^PMOL/L^210^70^Y^ D^RIA^^ ESTRADIOL (REFERENCE RANGE) PMOL/L^ ^ ^ D^RIA^^ ----------------------------------------^ ^ ^ D^RIA^^ FOLLICULAR : 0 - 921^ ^ ^ D^RIA^^ MID-CYCLE : 139 - 2382^ ^ ^ D^RIA^^ LUTEAL : 0 - 1145^ ^ ^ D^RIA^^ POSTMENOPAUSAL(NOT ON HRT) : 0 - 103^ ^ ^ D^RIA^^ POSTMENOPAUSAL( ON HRT ) : 0 - 528^ ^ ^ D^RIA^^ ----------------------------------------^ ^ ^ C^RIA^^^PROGESTERONE^N^^^NMOL/L^12^70^Y^ D^RIA^^ PROGESTERONE REF.RANGE NMOL/L^ ^ ^ D^RIA^^ -----------------------------^ ^ ^ D^RIA^^ FEMALE: ^ ^ ^ D^RIA^^ FOLLICULAR: 0 - 4^ ^ ^ D^RIA^^ LUTEAL: 4 - 51 ^ ^ ^ D^RIA^^ NOTE: VALUES GREATER THAN 16^ ^ ^ D^RIA^^ ARE STRONGLY SUGGESTIVE^ ^ ^ D^RIA^^ OF AN OVULATORY CYCLE.^ ^ ^ C^RIA^^^DHEA SULPHATE^A^^^UMOL/L^* 4.4 *^70^Y^ D^RIA^^ * REVIEW *^ ^ ^ D^RIA^^ * RESULT *^ ^ ^ D^RIA^^ DHEA-S REFERENCE RANGE (UMOL/L) ^ ^ ^ D^RIA^^ =================================^ ^ ^ D^RIA^^ AGE (YEARS) FEMALE MALE^ ^ ^ D^RIA^^ ---------------------------------^ ^ ^ D^RIA^^ NEWBORN 4.5-10.0 4.5-10.0^ ^ ^ D^RIA^^ 1-12 < 5.0 < 5.0^ ^ ^ D^RIA^^ 13-29 <11.0 <11.0^ ^ ^ D^RIA^^ 30-39 < 7.3 <14.0^ ^ ^ D^RIA^^ 40-49 < 6.5 <14.1^ ^ ^ D^RIA^^ 50-59 < 5.4 < 8.4^ ^ ^ D^RIA^^ 60-69 < 3.5 < 7.9^ ^ ^ D^RIA^^ 70-79 < 2.4 < 4.7^ ^ ^ D^RIA^^ ^ ^ ^ D^RIA^^ ^ ^ ^ D^RIA^^ ^ ^ ^ D^RIA^^ ^ ^ ^ B^XM60723^^20090401^LOULA^BIGGS^F^2525710386JV^19540824^F^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^416-528-2259^905-771-7771^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^127^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.378^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^6.2^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.38^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^86.2^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^29.1^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^337^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^13.6^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^214^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^3.8^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^1.7^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.4^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.3^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^HEMATOLOGY^^^hs-CRP^A^^^MG/L^17.6^70^Y^ D^HEMATOLOGY^^ CUT POINTS FOR CARDIAC RISK ASSESSMENT ARE:^ ^ ^ D^HEMATOLOGY^^ LOW RISK LEVEL <1.0 MG/L^ ^ ^ D^HEMATOLOGY^^ AVERAGE RISK LEVEL 1.0 - 3.0 MG/L^ ^ ^ D^HEMATOLOGY^^ HIGH RISK LEVEL >3.0 MG/L^ ^ ^ D^HEMATOLOGY^^ VALUES >8.0 MG/L INDICATE INFLAMMATORY^ ^ ^ D^HEMATOLOGY^^ CONDITIONS AND MAY NOT PREDICT CARDIAC RISK^ ^ ^ D^HEMATOLOGY^^ .^ ^ ^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^N^3.3^6.0^MMOL/L^5.6^70^N^ C^CHEMISTRY^^^CREATININE^N^60^115^UMOL/L^60^70^N^ C^CHEMISTRY^^^eGFR^N^^^^90^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^N^120^400^UMOL/L^301^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^145^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^3.9^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^108^70^N^ C^CHEMISTRY^^^TOTAL BILIRUBIN^N^0.0^23.0^UMOL/L^10.0^70^N^ C^CHEMISTRY^^^ALK PHOS^N^30^110^U/L^92^70^N^ C^CHEMISTRY^^^AST^N^6^42^U/L^16^70^N^ C^CHEMISTRY^^^CHOLESTEROL^N^TARGET <5.20^TARGET <5.20^MMOL/L^3.81^70^N^ C^CHEMISTRY^^^TRIGLYCERIDES^N^TARGET <1.71^TARGET <1.71^MMOL/L^1.57^70^N^ C^CHEMISTRY^^^HDL CHOLESTEROL^A^TARGET >1.29^TARGET >1.29^MMOL/L^1.00^70^N^ C^CHEMISTRY^^^LDL CHOLESTEROL^N^See Targets^See Targets^MMOL/L^2.10^70^N^ C^CHEMISTRY^^^CHOL/HDL RATIO^N^See Targets^See Targets^^3.81^70^Y^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ Clinical Risk Status: Target Lipid Levels^ ^ ^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ If Risk : LDL-C --Targets-- CHOL/HDL Ratio^ ^ ^ D^CHEMISTRY^^ Low : <5.1 and <6.0^ ^ ^ D^CHEMISTRY^^ Moderate : <3.5 and <5.0^ ^ ^ D^CHEMISTRY^^ High : <2.0 and <4.0^ ^ ^ D^CHEMISTRY^^ ...as per 2006 Canadian Guidelines^ ^ ^ C^RIA^^^TSH^N^0.30^4.70^MU/L^0.88^70^N^ C^RIA^^^T4 FREE^N^9.1^23.8^PMOL/L^15.7^70^N^ C^RIA^^^FREE T3^N^2.5^5.7^PMOL/L^4.5^70^N^ B^XS97153^^20090401^LINDA^HOW^F^7552086352RP^19680423^P^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^416-269-5484^905-771-7771^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^136^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.408^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^10.4^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.13^70^N^ C^HEMATOLOGY^^^MCV^A^80^97^FL^98.6^70^N^ C^HEMATOLOGY^^^MCH^A^27.0^32.0^PG^32.9^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^334^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^13.3^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^235^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^6.8^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.5^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.8^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.2^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.1^70^N^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^A^3.3^6.0^MMOL/L^6.9^70^N^ C^CHEMISTRY^^^CREATININE^A^60^115^UMOL/L^56^70^N^ C^CHEMISTRY^^^eGFR^N^^^^103^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^A^120^400^UMOL/L^440^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^140^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^3.5^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^98^70^N^ C^CHEMISTRY^^^TOTAL BILIRUBIN^N^0.0^23.0^UMOL/L^14.0^70^N^ C^CHEMISTRY^^^ALK PHOS^N^30^110^U/L^77^70^N^ C^CHEMISTRY^^^AST^A^6^42^U/L^67^70^N^ C^CHEMISTRY^^^CHOLESTEROL^N^TARGET <5.20^TARGET <5.20^MMOL/L^4.24^70^N^ C^CHEMISTRY^^^TRIGLYCERIDES^A^TARGET <1.71^TARGET <1.71^MMOL/L^2.08^70^N^ C^CHEMISTRY^^^HDL CHOLESTEROL^A^TARGET >1.29^TARGET >1.29^MMOL/L^1.01^70^N^ C^CHEMISTRY^^^LDL CHOLESTEROL^N^See Targets^See Targets^MMOL/L^2.28^70^N^ C^CHEMISTRY^^^CHOL/HDL RATIO^A^See Targets^See Targets^^4.20^70^Y^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ Clinical Risk Status: Target Lipid Levels^ ^ ^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ If Risk : LDL-C --Targets-- CHOL/HDL Ratio^ ^ ^ D^CHEMISTRY^^ Low : <5.1 and <6.0^ ^ ^ D^CHEMISTRY^^ Moderate : <3.5 and <5.0^ ^ ^ D^CHEMISTRY^^ High : <2.0 and <4.0^ ^ ^ D^CHEMISTRY^^ ...as per 2006 Canadian Guidelines^ ^ ^ B^YH03548^^20090331^JOYCE^PONY^F^7332205413LE^19550326^F^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^905-502-9611^905-771-7771^31-MAR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^121^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^A^0.37^0.47^L/L^0.360^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^6.0^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.00^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^89.9^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^30.2^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^336^70^N^ C^HEMATOLOGY^^^RDW^A^11.0^14.5^%^15.6^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^268^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^3.6^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.0^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.4^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.0^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^HEMATOLOGY^^^RHEUMATOID FACTOR^A^BELOW 20^BELOW 20^KU/L^257^70^N^ C^HEMATOLOGY^^^hs-CRP^A^^^MG/L^1.9^70^Y^ D^HEMATOLOGY^^ CUT POINTS FOR CARDIAC RISK ASSESSMENT ARE:^ ^ ^ D^HEMATOLOGY^^ LOW RISK LEVEL <1.0 MG/L^ ^ ^ D^HEMATOLOGY^^ AVERAGE RISK LEVEL 1.0 - 3.0 MG/L^ ^ ^ D^HEMATOLOGY^^ HIGH RISK LEVEL >3.0 MG/L^ ^ ^ D^HEMATOLOGY^^ VALUES >8.0 MG/L INDICATE INFLAMMATORY^ ^ ^ D^HEMATOLOGY^^ CONDITIONS AND MAY NOT PREDICT CARDIAC RISK^ ^ ^ D^HEMATOLOGY^^ .^ ^ ^ C^HEMATOLOGY^^^ANA^N^NEGATIVE^NEGATIVE^^NEGATIVE^70^N^ C^HEMATOLOGY^^^READING DOCTOR^N^^^^^70^Y^ D^HEMATOLOGY^^ A. SEIDENFELD, M.D.,F.R.C.P.(C)^ ^ ^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^N^3.3^6.0^MMOL/L^5.5^70^N^ C^CHEMISTRY^^^CREATININE^N^60^115^UMOL/L^68^70^N^ C^CHEMISTRY^^^eGFR^N^^^^78^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^N^120^400^UMOL/L^363^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^142^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^4.5^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^104^70^N^ C^CHEMISTRY^^^TOTAL BILIRUBIN^N^0.0^23.0^UMOL/L^12.0^70^N^ C^CHEMISTRY^^^ALK PHOS^N^30^110^U/L^101^70^N^ C^CHEMISTRY^^^AST^N^6^42^U/L^31^70^N^ C^CHEMISTRY^^^C3^A^0.70^1.76^G/L^1.82^70^N^ C^CHEMISTRY^^^C4^N^0.16^0.45^G/L^0.42^70^N^ C^CHEMISTRY^^^CHOLESTEROL^A^TARGET <5.20^TARGET <5.20^MMOL/L^5.57^70^N^ C^CHEMISTRY^^^TRIGLYCERIDES^A^TARGET <1.71^TARGET <1.71^MMOL/L^3.81^70^N^ C^CHEMISTRY^^^HDL CHOLESTEROL^A^TARGET >1.29^TARGET >1.29^MMOL/L^1.19^70^N^ C^CHEMISTRY^^^LDL CHOLESTEROL^A^See Targets^See Targets^MMOL/L^2.65^70^N^ C^CHEMISTRY^^^CHOL/HDL RATIO^A^See Targets^See Targets^^4.68^70^Y^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ Clinical Risk Status: Target Lipid Levels^ ^ ^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ If Risk : LDL-C --Targets-- CHOL/HDL Ratio^ ^ ^ D^CHEMISTRY^^ Low : <5.1 and <6.0^ ^ ^ D^CHEMISTRY^^ Moderate : <3.5 and <5.0^ ^ ^ D^CHEMISTRY^^ High : <2.0 and <4.0^ ^ ^ D^CHEMISTRY^^ ...as per 2006 Canadian Guidelines^ ^ ^ C^RIA^^^TSH^N^0.30^4.70^MU/L^0.63^70^N^ C^RIA^^^T4 FREE^N^9.1^23.8^PMOL/L^9.4^70^N^ C^RIA^^^FREE T3^N^2.5^5.7^PMOL/L^4.3^70^N^ B^YX16191^^20090401^MARIA^BIGS^F^3429942141LA^19620304^F^0696170^DR. JEFFREY M. BROWN^SUITE 214^7330 YONGE STREET^THORNHILL, ONTARIO L4J 1V8^^1475^^^905-790-9822^905-771-7771^01-APR-09^ C^HEMATOLOGY^^^HEMOGLOBIN^N^115^165^G/L^136^70^N^ C^HEMATOLOGY^^^HEMATOCRIT^N^0.37^0.47^L/L^0.407^70^N^ C^HEMATOLOGY^^^WBC COUNT^N^4.0^11.0^X10 9/L^6.1^70^N^ C^HEMATOLOGY^^^RBC COUNT^N^3.80^5.80^X10 12/L^4.54^70^N^ C^HEMATOLOGY^^^MCV^N^80^97^FL^89.5^70^N^ C^HEMATOLOGY^^^MCH^N^27.0^32.0^PG^29.9^70^N^ C^HEMATOLOGY^^^MCHC^N^320^360^G/L^334^70^N^ C^HEMATOLOGY^^^RDW^N^11.0^14.5^%^14.2^70^N^ C^HEMATOLOGY^^^PLATELET COUNT^N^150^400^X10 9/L^239^70^N^ C^HEMATOLOGY^^^ABSOLUTE: NEUTROS^N^2.0^7.5^X10 9/L^3.5^70^N^ C^HEMATOLOGY^^^(A) LYMPH^N^1.1^3.3^X10 9/L^2.1^70^N^ C^HEMATOLOGY^^^(A) MONO^N^0.0^0.8^X10 9/L^0.4^70^N^ C^HEMATOLOGY^^^(A) EOS^N^0.0^0.5^X10 9/L^0.1^70^N^ C^HEMATOLOGY^^^(A) BASO^N^0.0^0.2^X10 9/L^0.0^70^N^ C^CHEMISTRY^^^GLUCOSE FASTING-SER^A^3.3^6.0^MMOL/L^8.4^70^N^ C^CHEMISTRY^^^UREA^N^3.0^8.2^MMOL/L^4.4^70^N^ C^CHEMISTRY^^^CREATININE^N^60^115^UMOL/L^66^70^N^ C^CHEMISTRY^^^eGFR^N^^^^83^70^Y^ D^CHEMISTRY^^ **** **** ****^ ^ ^ D^CHEMISTRY^^ For patients of African descent, the reported^ ^ ^ D^CHEMISTRY^^ eGFR must be multiplied by a correction^ ^ ^ D^CHEMISTRY^^ factor of 1.21.^ ^ ^ D^CHEMISTRY^^ **** **** ****^ ^ ^ C^CHEMISTRY^^^URATE^N^120^400^UMOL/L^273^70^N^ C^CHEMISTRY^^^SODIUM^N^135^146^MMOL/L^141^70^N^ C^CHEMISTRY^^^POTASSIUM^N^3.5^5.2^MMOL/L^4.0^70^N^ C^CHEMISTRY^^^CHLORIDE^N^95^108^MMOL/L^106^70^N^ C^CHEMISTRY^^^TOTAL BILIRUBIN^N^0.0^23.0^UMOL/L^11.0^70^N^ C^CHEMISTRY^^^ALK PHOS^N^30^110^U/L^81^70^N^ C^CHEMISTRY^^^AST^A^6^42^U/L^88^70^N^ C^CHEMISTRY^^^HEMOGLOBIN A1C^A^0.040^0.060^^0.075^70^N^ C^CHEMISTRY^^^CHOLESTEROL^N^TARGET <5.20^TARGET <5.20^MMOL/L^4.28^70^N^ C^CHEMISTRY^^^TRIGLYCERIDES^N^TARGET <1.71^TARGET <1.71^MMOL/L^0.80^70^N^ C^CHEMISTRY^^^HDL CHOLESTEROL^A^TARGET >1.29^TARGET >1.29^MMOL/L^0.99^70^N^ C^CHEMISTRY^^^LDL CHOLESTEROL^A^See Targets^See Targets^MMOL/L^2.93^70^N^ C^CHEMISTRY^^^CHOL/HDL RATIO^A^See Targets^See Targets^^4.32^70^Y^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ Clinical Risk Status: Target Lipid Levels^ ^ ^ D^CHEMISTRY^^ *----------*----------*----------*----------*^ ^ ^ D^CHEMISTRY^^ If Risk : LDL-C --Targets-- CHOL/HDL Ratio^ ^ ^ D^CHEMISTRY^^ Low : <5.1 and <6.0^ ^ ^ D^CHEMISTRY^^ Moderate : <3.5 and <5.0^ ^ ^ D^CHEMISTRY^^ High : <2.0 and <4.0^ ^ ^ D^CHEMISTRY^^ ...as per 2006 Canadian Guidelines^ ^ ^ C^RIA^^^TSH^N^0.30^4.70^MU/L^0.86^70^N^ C^RIA^^^T4 FREE^N^9.1^23.8^PMOL/L^15.4^70^N^ C^RIA^^^FREE T3^N^2.5^5.7^PMOL/L^4.7^70^N^
require('stringutil')
require('node')
require('dateparser')
function main(Data)
local R = ProcessLab(Data)
end
function node.D(Data)
local T = dateparse.parse(Data:nodeValue())
return os.date('%Y-%m-%d %H:%M:%S', T)
end
function ProcessLab(Data)
local D = hl7.parse{vmd='lab.vmd', data=Data}
local Out = db.tables{vmd='lab.vmd', name='Labs'}
print(#D.Patient..' patients in batch.')
for i=1, #D.Patient do
MapPatient(Out.Patient[i], D.Patient[i].B)
end
return Out
end
function MapPatient(T, I)
T.FirstName = I[4]:nodeValue():capitalize()
T.LastName = I[5]:nodeValue():capitalize()
T.Dob = I[3]:D()
end
What the bare bones of the Translator gives you [top]
If you try and map an incorrectly formatted date time string to a date/time column you are likely to see an error something like this:
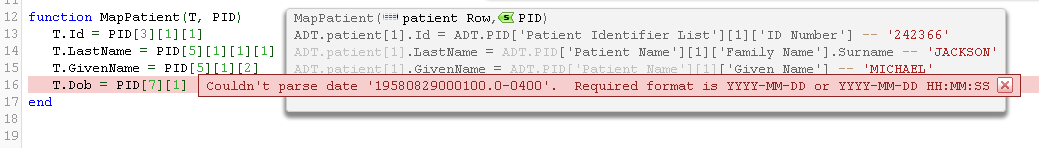
It’s not hard to write string munching code to break apart that date/time format into pieces and format it in the desired form. It’s easier to use the fuzzy date time parser we wrote though have it handle the conversion automatically.
Using the fuzzy date/time parser [top]
The minimal set of steps required is:
- Import the channel the Parse any date time value channel from the Builtin: Iguana Date/Time repository.
- This will the shared date.parse module which you can re-use in your channel
- Add the code
local dateparse = require 'date.parse'at the top of the main module.
Note: The code is show as require(‘dateparse’) in this article but that is the old way and has been superseded. - Then you use can access the parser using the “D” method of any node object like this:
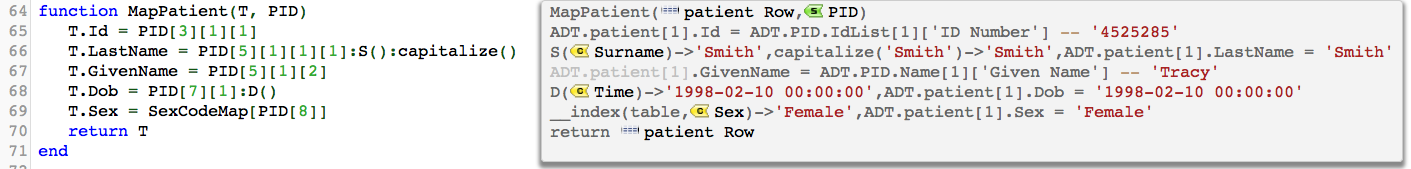
The D method is a function existing in the node table. See methods in Lua.
For the most part this will just work unless you come across date/time formats that the fuzzy date/time parser has not be configured to understand.
Ambiguous Date/Time Formats [top]
Some date time formats can be ambiguous. For instance consider 07-Sep-06. That could mean the 7th of September 2006 or 6th September 2007.
It’s bad form that such formats exist in interchange formats, but they do. When you encounter such situations you can explicitly pass a format parameter to the D() function giving the expected format of the date/time value. For instance:
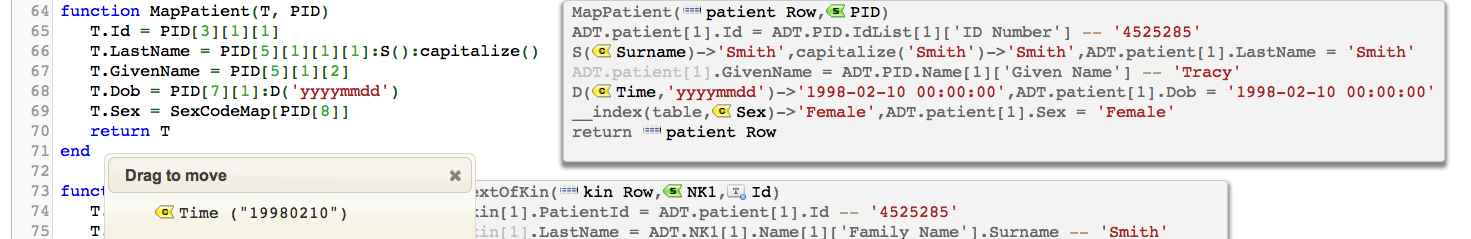
In this case we give a trivial example of passing in ‘yyyymmdd’.
Fortunately when you do get such formats you can leverage the sample data and annotations within the environment to quickly determine if you have made the correct assumptions.
The formatting parameters that the parser knows are covered next.
Complete documentation for the fuzzy date/time parser [top]
If you look at the source code for the parser you’ll see that the node.D() function invokes the the string.D() function wich invokes the actual implementation which is called dateparse.parse().
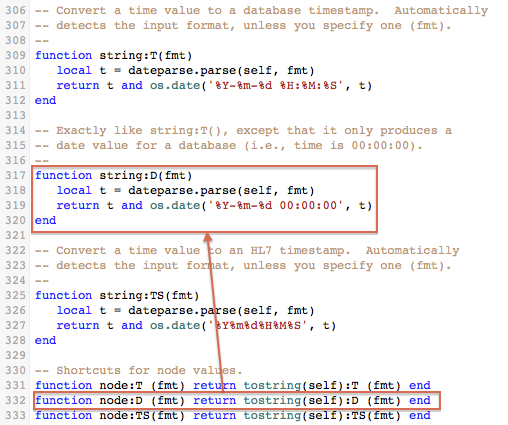
The dateparse.parse() function recognizes the format, and returns two values: a Lua time value (t), and a table with the date/time components (d). The time value (t) can be compared with the Lua routine os.difftime() or formatted as text with os.date(). If you want to handle add and compare date/times then you probably will need to use this interface.

The component table (d) has various fields usable with Lua routines (i.e., year, month, day, hour, min, sec and isdst), as well as fields for partial seconds (sec_fraction) and time zones (tz and tz_offset). As you can see in the example, we can compute the point in time exactly fourteen days later just by adding 14 to d.day. Modifying the table (d) does not affect the time value (t), of course, but you can use os.time() to compute the new future timestamp.
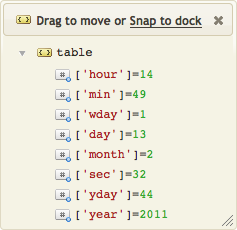
Now that we have our new time value (t), we use os.date() to format it HL7-style.
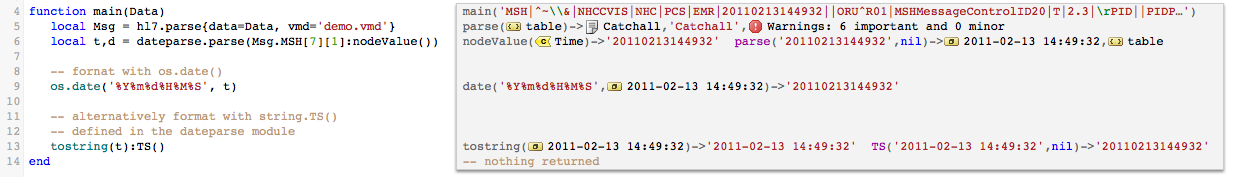
If wanted to format it differently, say in an XML format we could either:
- Make a new method like function node.XD that would use this new format.
- Implement a new copy of function node.D in the main module to override the implementation in dateparse.lua.
- Edit the dateparse.lua module itself. Not recommended, but it is your code.
It’s not all Roses
Take care when testing the fuzzy parser, as sometimes it gets it wrong. Suppose a message had 01/10/07 or 07-Oct-01, for October 1st, 2007. The fuzzy parser would get both wrong, interpreting them as January 10th, 2007, and October 7th, 2001, respectively.
It tries to match common formats, but there are a ton of ways to write dates out, so it can’t always get it right. You can always specify the format (described below), but you have the source of the module: make it your own! We tried to make it easy to read and modify, so skim it for the list of known formats and edit them to your heart’s content.
Custom Formats
The dateparse.parse() function is pretty smart, but it can’t predict everything. Suppose you get a more free-form date/time in a message, like “2PM March 5th, ’77”. Currently dateparse.parse() doesn’t understand that format. The module contains a list of known formats, which you can add to. Alternatively, you can include the format details in the call to dateparse.parse() itself, as in the following example.
require 'dateparse'
require 'node'
function main(Data)
local Ts,Tm = dateparse.parse(
"2PM March 5th '77",
"H[:MM]tt mmmm dw [']yy[yy]")
tostring(t):TS() --> 19770305140000
end
This is an excellent approach when you encounter very odd date/time formats, and you don’t expect them to be found elsewhere. No need to complicate the dateparse module for one-off problems.
In the format we used: H will match the one-digit or two-digit hour; [:MM] will match a colon and a two-digit minute, if present (the brackets make this whole part optional); and tt matches AM or PM. For the date part, mmmm matches the month name (full or abbreviated), d matches the day (one-digit or two-digit), w matches the “th” (actually it will match and ignore any word); and [‘]yy[yy] will match a two-digit or four-digit year, possibly preceded by an apostrophe.
Obviously this example pattern matches more than the one example date/time we are working with, but with hand-written date/times, it’s best to be flexible. Any format you specify must match the entire date/time string you supply. If you expect any extra words, numbers or punctuation, you need to specify them in the pattern—just make them [optional]. The full pattern language is detailed after the module below.
Time Zones
The dateparse.parse() function returns two values: a Lua time value (t) and a date/time component table (d). Time zone information is understood by the parser, but not by Lua. The Lua time value (t) returned will not be adjusted when time zone information is present. Instead we add a couple fields to the component table (d), so that you can adjust the time value yourself, if you so desire.
When a time zone abbreviation (e.g., EST) is recognized in a date/time string, d.tz_offset is given the offset in minutes from Universal Time Coordinated (UTC), and the abbreviation is stored in d.tz (in uppercase). E.g., with “EST”, d.tz_offset would be set to -300 (EST is UTC-05:00).
When an offset itself is present as (+/-)HH:MM (or without the colon, as in HL7 timestamps), the d.tz_offset field is computed given this value, and d.tz is set to “UTC(+/-)HH:MM” (always with a colon). E.g., if the offset -0500 were read in an HL7 timestamp, d.tz and d.tz_offset would be set to “UTC-05:00” and -300, respectively.
When writing your own formats, “zzzz” can be used to match time zone offsets (including the +/- part), and “ZZZ” can be used to match abbreviations. See the note on formats below for details.
Partial Seconds
Some timestamps can be incredibly accurate. In an HL7 timestamp, for instance, you can specify values accurate to 1/10th of a millisecond (four decimal places). As with time zones, Lua time values cannot contain this information, but we provide it in the d.sec_fraction field in the example above. E.g., with the HL7 timestamp “19771020213003.1415-0500”, the d.sec_fraction value would be 0.1415.
When writing your own formats, “ssss” can be used to match partial second values (it will not match against decimal point). E.g., “HHMMSS[.ssss]”.
Date/time Formats
The following table lists the patterns allowed in date/time format strings. Note that unless your format contains spaces, spaces in date/time strings won’t be accepted by the parser. E.g., the HL7 format “yyyymmddHHMM” will not match “1977 10 20 21 30” because it contains spaces not found in the pattern.
Note, however, that having spaces (or commas) in your pattern does not mean that they are required. E.g., “mmmm dw, yyyy” will still match “October20th1977”. This allows for more flexibility, without more effort from you. When spaces are really required, as in patterns like “yyyy/m/d H:MM” (between the d and the H), the absence of spaces will make the pattern fail to match. For a value like “1977/1/115:30”, even a human would have to guess if the hour is 15 or if the day is 11.
| Pattern | Matches What? |
|---|---|
| yy | Year, two digits. With ’69–99 in the 1900s. |
| yyyy | Year, four digits. |
| m | Month, one or more digits. |
| mm | Month, two digits. |
| mmm | Month, three character abbreviation. |
| mmmm | Month, abbrev. or full name. |
| d | Day, one or more digits. |
| dd | Day, two digits. |
| ddd | Week-day, three character abbreviation (ignored) |
| dddd | Week-day, abbrev. or full name (ignored) |
| H | Hour, one or more digits. |
| HH | Hour, two digits. |
| M | Minute, one or more digits. |
| MM | Minute, two digits. |
| S | Second, one or more digits. |
| SS | Second, two digits. |
| ssss | Second fraction, one or more digits (e.g., “SS[.ssss]”). |
| t | “A” or “P” (for AM and PM, respectively). |
| tt | “AM” or “PM” |
| zzzz | Time zone offset, e.g., “-05:00” (colon optional). |
| ZZZ | Time zone abbreviation, one word. |
| A Space | Any amount of whitespace (including none). |
| , | A comma (optional; may be preceded by whitespace). |
| w | Any word (ignored) |
| n | Any number (ignored; -/+ are not matched) |
| (other*) | *Except letters. Matched exactly as-is. |
The dateparse Module
The source code for the fuzzy date/time matcher can be downloaded from the Date/Time repository.
Using os.date to output arbitrary date/time formats [top]
The os.date function is great for outputting arbitrary date/time formats. It’s straightforward to use, here is an example in this case formatting the current time:

Here is another example, in this case we parse the input time using the fuzzy date parser and then output it using os.date:
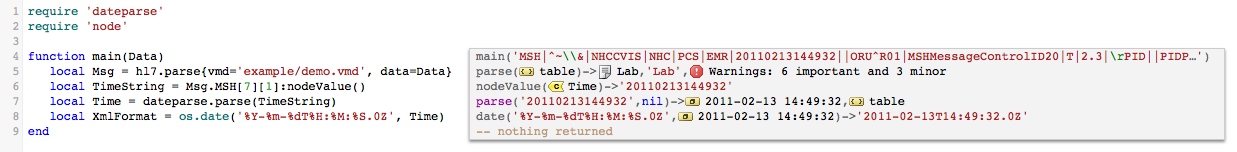
You can see how flexible os.date is – especially if you mix it in with using the Lua string library for doing arbitrary string handling.
The formatting codes accepted within the date formatting string are the same as those supported by strftime. This table shows the available codes:
| Code | Replaced by | Example |
|---|---|---|
| %a | Abbreviated weekday name | Thu |
| %A | Full weekday name | Thursday |
| %b | Abbreviated month name | Aug |
| %B | Full month name | August |
| %c | Date and time representation | Thu Aug 23 14:55:02 2001 |
| %d | Day of the month (01-31) | 23 |
| %H | Hour in 24h format (00-23) | 14 |
| %I | Hour in 12h format (01-12) | 02 |
| %j | Day of the year (001-366) | 235 |
| %m | Month as a decimal number (01-12) | 08 |
| %M | Minute (00-59) | 55 |
| %p | AM or PM designation | PM |
| %S | Second (00-61) | 02 |
| %U | Week number with the first Sunday as the first day of week one (00-53) | 33 |
| %w | Weekday as a decimal number with Sunday as 0 (0-6) | 4 |
| %W | Week number with the first Monday as the first day of week one (00-53) | 34 |
| %x | Date representation * | 08/23/01 |
| %X | Time representation * | 14:55:02 |
| %y | Year, last two digits (00-99) | 01 |
| %Y | Year | 2001 |
| %% | A % sign | % |
One useful trick for getting the date/time in a useful format is to use this:
local T = os.date("*t")
This returns a Lua table of date/time components, i.e.:
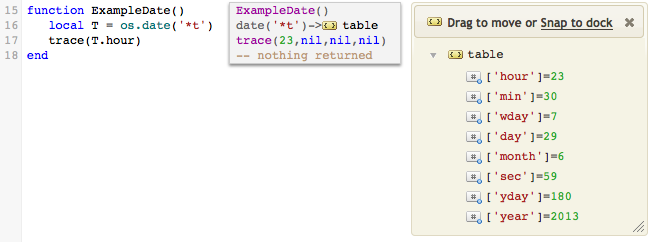
For more documentation on os.date and os.time see the Lua reference manual online.
The following code snippet is handy in exercising all the different available formatting codes:
function CurrentTime() local F = [[%%a=%a %%A=%A %%b=%b %%B=%B %%c=%c %%d=%d %%H=%H %%I=%I %%j=%j %%m=%m %%M=%M %%p=%p %%S=%S %%U=%U %%w=%w %%W=%W %%x=%x %%X=%X %%y=%y %%Y=%Y ]] return os.date(F, os.time()) end
Don’t be afraid to mix in a bit of string parsing [top]
There are going to be times when you need to parse or generate odd date formats which are not supported out of the box by the fuzzy date parser or os.date. In these situations it is good to remember that there is nothing wrong with whipping in and doing a bit of arbitrary string manipulation. The visual nature of the Translator excels at making this easy. There really is no limit to what you can do with the Translator.
For example an example of this check out Lev’s tip with parsing an odd date.
An elegant way to do this processing is to wrap the code up in a custom extension method. That makes it easy to use.
Some user suggestions for other useful date/time libraries [top]
Some of our users occasionally find libraries that they recommend to us. One library that Dominic Green found useful was this date-time manipulation library. If you find a useful library you think other people would like let us know.
